 |
 |
||||
 |
|||||
Research
1) Molecular mechanism of retrotransposition of LINE and its application
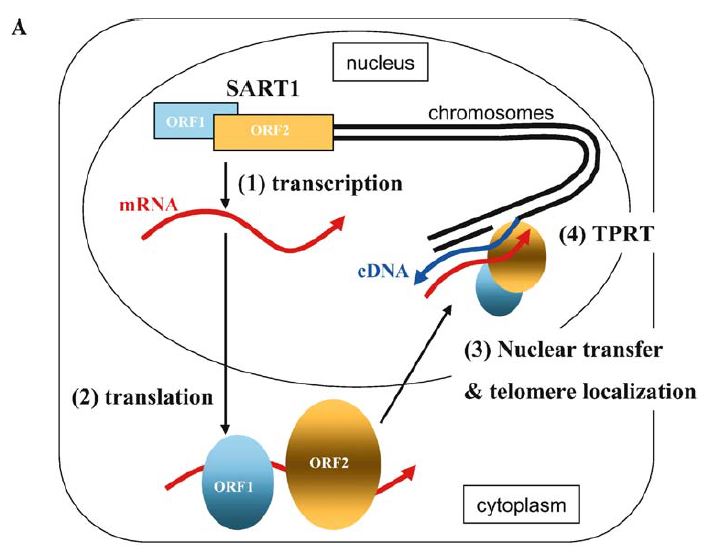
LINE occupies over 20% of the human genome and cause many genetic diseases, but retrotransposition mechanisms remain to be elucidated. We have studied target-specific LINE in insects and recently established the retrotransposition system with baculovirus-mediated gene delivery. Using this system, we continue to clarify the essential domains for LINE retrotransposition mechanisms. We are also applying this system to sequence-specific gene delivery and gene therapy vector.
Important papers are marked by yellow.
1) Fujiwara, H., Ogura, T., Takada, N., Miyajima, N., Ishikawa, H. and Maekawa, H.: Introns and their flanking sequences of Bombyx mori rDNA. Nucleic Acids Res. (1984) 12, 6861-6869.
2) Fujiwara, H., Ninaki, O., Kobayashi, M., Kusuda, J. and Maekawa, H.: Chromosomal fragment responsible for genetic mosaicism in larval body marking of the silkworm, Bombyx mori. Genet. Res. (1991) 57, 11-16.
3) Okazaki, S., Tsuchida, K., Maekawa, H., Ishikawa, H. and Fujiwara, H.: Identification of a pentanucleotide telomeric sequence, (TTAGG)n, in the silkworm, Bombyx mori and in other insects. Mol. Cell. Biol. (1993) 13, 1424-1432.
4) Fujiwara, H., Yanagawa, M. and Ishikawa, H.: Mosaic formation by developmental loss of chromosomal fragment in a genetic mosaic, mottled striped of the silkworm, Bombyx mori. Roux's Arch. Dev. Biol. (1994) 203, 389-396.
5) Fujiwara, H. and Maekawa, H.: RFLP analysis of chromosomal fragments in mottled striped strains of the silkworm, Bombyx mori. Chromosoma (1994) 103, 468-474.
6) Okazaki, S., Ishikawa, H. and Fujiwara, H: Structural analysis of TRAS1, a novel family of telomeric repeat associated retrotransposons in the silkworm, Bombyx mori. Mol. Cell. Biol. (1995) 15, 4545-4552.
7) Takahashi, H., Okazaki, S. and Fujiwara, H.: A new family of site-specific retrotransposons, SART1, is inserted into telomeric repeats of the silkworm, Bombyx mori.. Nucleic Acids Res. (1997) 25, 1578-1584.
8) Takahashi, H. and Fujiwara, H.: Transcription analysis of the telomeric repeat-specific retrotransposons TRAS1 and SART1 of the silkworm Bombyx mori. Nucleic Acids Res. (1999) 27, 2015-2021.
9) Sasaki, T. and Fujiwara, H.: Detection and distribution patterns of telomerase activity in insects. Eur. J. Biochem. (2000) 267, 3025-3031.
10) Fujiwara, H., Nakazato, Y., Okazaki, S. and Ninaki, O.: Stability and telomere structure of chromosomal fragments in two different mosaic strains of the silkworm, Bombyx mori. Zool Sci. (2000) 17, 743-750.
11) Anzai, T., Takahashi, H. and Fujiwara, H.: Sequence-specific recognition and cleavage of telomeric repeat (TTAGG)n by endonuclease of non-LTR retrotransposon, TRAS1. Mol Cell. Biol. (2001) 21, 100-108.
12) Kubo, Y., Okazaki, S., Anzai, T. and Fujiwara, H.: Structural and phylogenetic analysis of TRAS, telomeric repeat-specific non-LTR retrotransposon families in lepidopteran insects. Mol. Biol. Evol, (2001) 18. 848-857.
13) Kojima, K, Kubo, Y. and Fujiwara, H.: Complex and tandem repeat structure in sub-telomere of crickets, T. taiwanemma. J. Mol. Evol, (2002) 54, 474-485.
14) Takahashi, H. and Fujiwara, H.: Transplantation of target site specificity by swapping endonuclease domains of two LINEs. EMBO. J. (2002) 21, 408-417.
15) Kojima, K. K. and Fujiwara, H.: Evolution of target specificity in R1 clade non-LTR retrotransposons. Mol. Biol. Evol, (2003) 20, 351-361.
16) Kojima, K. K. and Fujiwara, H.: Cross-genome screening of novel sequence-specific non-LTR retrotransposons: various multicopy RNA genes and microsatellites are selected as targets. Mol. Biol. Evol. (2004) 21, 207-217.
17) Matsumoto, T., Takahashi, H. and Fujiwara, H. Targeted nuclear import of ORF1 protein is required for in vivo retrotransposition of telomere-specific non-long terminal repeat retrotransposon, SART1. Mol. Cell Biol. (2004) 24, 105-122.
18) Osanai, M., Takahashi, H., Kojima, K. K., Hamada, M. and Fujiwara, H.: Novel motifs in 3’-untranslated region required for precise reverse transcription start of telomere specific LINE, SART1. Mol. Cell Biol. (2004) 24, 7902-7913.
19) Maita, N., Anzai, T., Aoyagi, H., Mizuno, H. and Fujiwara, H.: Crystal structure of the endonuclease domain encoded by the telomere-specific LINE, TRAS1. J. Biol. Chem (2004) 279, 41067-41076.
20) Anzai T., Osanai, M., Hamada, M. and Fujiwara, H.: Functional roles of read-through 28S rRNA sequence in in vivo retrotransposition of non-LTR retrotransposon, R1Bm. Nucleic Acids Res. (2005) 33, 1993-2002.
21) Kojima, K. K. and Fujiwara, H.: An extraordinary retrotransposon family encoding dual endonucleases, Genome Research, in press (2005)
22) Matsumoto, T. Hamada, M. Osanai, M. and Fujiwara, H.: Essential domains for Ribonucleoprotein complex formation required for retrotransposition of a telomere specific non-LTR retrotransposon SART1. Mol. Cell. Biol. 26, 5168-5179 (2006)
23) Ando, T., Fujiyuki, T., Kawashima, T., Morioka, M., Kubo, T. and Fujiwara, H.: In vivo gene transfer into the honeybee using a nucleopolyhedrovirus vector. Biochem. Biophys. Res. Comm. 352, 335-340 (2007)
24) Kawashima, T., Osanai, M., Futahashi, R., Kojima, T. and Fujiwara, H.: A novel target-specific gene delivery system combining baculovirus and sequence-specific LINEs. Virus Research. 127, 49-60 (2007)
25) Maita, N., Aoyagi, H, Osanai, M., Shirakawa, M. and Fujiwara, H.: Characterization of the sequence specificity of the R1Bm endonuclease domain by structural and biochemical studies. Nucleic Acids Research, 35, 3918-3927 (2007)
26) Osanai-Futahashi, M. and Fujiwara, H.: Coevolution of telomeric repeats and telomeric-repeat-specific non-LTR retrotransposons in insects. Molecular Biology and Evolution. 28, 2983-2986 (2011)
27) Fujiwara, H.: Site-specific non-LTR retrotransposons. Chapter 50 in “Mobile DNA III” eds. by N. Craig et al., pp1147-1163. doi: 10.1128/9781555819217 (2015)
28) Fujiwara, H.: Site-specific non-LTR retrotransposons. Microbiology Spectrum, 3(2). April doi: 10.1128/microbiolspec.MDNA3-0001-2014 (2015)
29) Nichuguti Narisu, Hayase M. and Fujiwara, H.: Both exact target site sequence and long poly(A) tail are required for precise insertion of the 18S rDNA-specific non-long terminal repeat retrotransposon R7Ag. Mol. Cell. Biol36, 1494-1508. (2016)
Back to home
|
|
|
| All Rights Reserved, Copyright(C), Laboratory of Innovational Biology, Department of Integrated Biosciences, Graduate School of Frontier Sciences, The University of Tokyo |